Scientists discover the evolutionary secret behind different animal life cycles
Researchers show a link between the time of activation of dozens of genes in the embryo and how the life cycles of animals evolved.
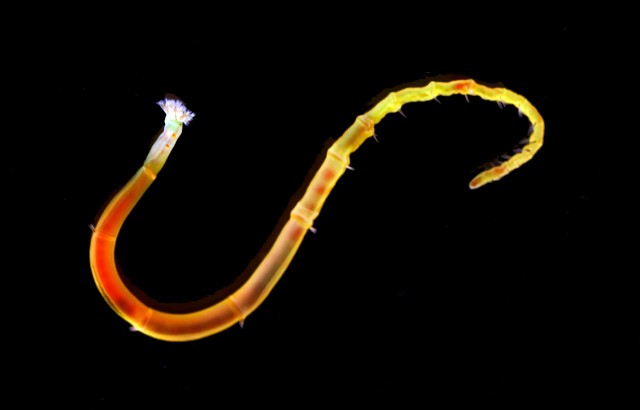
The worm for which the researchers sequenced the genome
For over 100 years, biologists have wondered why animals display different types of life cycles. Some species, like us humans and most vertebrates, develop directly into a fully formed—yet smaller—version of an adult. In contrast, many other animals give rise to beautifully diverse intermediate forms we call larvae, which then metamorphose into the adult.
Still, the understanding researchers had of why larvae exist and how they originated was limited. More importantly, large-scale comparative studies addressing this issue had not previously used modern techniques based on sequencing the genetic information of an animal—the genome—and finding how the organism uses this information while growing. Until now.
In a study led by a team at Queen Mary University of London (QMUL), published in the prestigious journal Nature, researchers uncover for the first time the mechanism that likely explains how embryos form either a larva or a miniature version of the adult.
In their paper, they prove that the timing of activation of essential genes involved in embryogenesis—the transformation of a fertilised egg into an organism—correlates with the presence or absence of a larval stage and with whether the larva feeds from their surroundings or relies on nourishment the mother deposited in the egg.
Francisco M. Martín-Zamora, PhD candidate at Queen Mary and co-first author of the study, said: “It’s impressive to see how evolution shaped the way animal embryos “tell the time” to activate important groups of genes earlier or later in development. Suppose a larval stage is no longer essential for your survival. In that case, it might be evolutionarily advantageous to, for example, activate the genes to form the trunk earlier and develop straight into an adult instead.”
This new study used state-of-the-art approaches to decode the genetic information, activity, and regulation in three species of marine invertebrate worms called annelids. They combined these with public datasets from other species in a large-scale study involving over 600 datasets of more than 60 species separated by more than 500 million years of evolution. “Only by combining experimental datasets generated in the lab and systematic computational analyses were we able to unravel this new undiscovered biology”, said Dr Ferdinand Marlétaz, a main collaborator of the study from University College London.
Dr Yan Liang, postdoctoral researcher from Queen Mary and co-first author of the work, said: “While the techniques had been around for some years, no team had used them for this purpose. The datasets we generated and the methodologies we developed will be tremendously powerful resources for other researchers.”
Dr Chema Martín-Durán, Senior Lecturer in Organismal Biology at Queen Mary and the senior author in this research, said: “Developmental biology largely focuses on mice, flies and other well-established species we know as model organisms. Our study demonstrates that the fascinating biology of the often-overlooked non-model species is critical to understand how animal development works and how it evolved.”
Genes involved in forming the trunk—the body region that follows the head and runs until the tail—are paramount. Some species will form larvae with virtually no trunk, known as “head larvae”, and might have been present as far back as in the ancestor of all animals with head and tail. Direct development and forming a small adult straight from embryogenesis would have evolved later in many animal groups, like us and most vertebrates, as genes to form the trunk get activated earlier in embryogenesis, and larval traits are progressively lost.
“We are hopeful that other researchers in the field will continue studying the exciting topic of the evolution of animal life cycles and provide further evidence for the hypothesis we put forward.”, Dr Andreas Hejnol said, Professor at the Friedrich-Schiller-University Jena, Germany, and collaborator of the team.
Though led by QMUL researchers, the present work is a multidisciplinary collaboration of over a dozen researchers, with collaborators from University College London, Imperial College London, and the National Museum Wales, in the UK; the Okinawa Institute for Science and Technology in Japan; the Friedrich-Schiller-University Jena, in Germany; and the University of Bergen, in Norway.
The European Union Horizon 2020 programme from the European Research Council (ERC) funded the work, as well as the Biotechnology and Biological Sciences Research Council (BBSRC), the Royal Society, and the Japan Society for the Promotion of Science.
Nature paper 'Annelid functional genomics reveal the origins of bilaterian life cycles'
Related items

9 December 2024

28 November 2024
