Biomolecular Simulations
Biomolecular Modelling
Research in this section focuses on the computational study of protein dynamics and its relationship to protein function. Systems currently under investigation range from muscle proteins to GPCR receptors and to kinases. The section is also involved in the development of general methods for the modelling of conformational transitions in proteins and of protein-ligand interactions.
Techniques: Molecular Dynamics simulations of biomolecules, Homology Modelling, Molecular Docking, Virtual Screening.
Keywords: protein dynamics, muscle proteins, myosin modulators, in silico drug design, binding pocket dynamics.
Recent Publications
Motta S, Pandini A, Fornili A, Bonati L (2021). Reconstruction of ARNT PAS-B Unfolding Pathways by Steered Molecular Dynamics and Artificial Neural Networks. Journal of Chemical Theory and Computation
10.1021/acs.jctc.0c01308
https://qmro.qmul.ac.uk/xmlui/handle/123456789/72283
Fornili A, Hashem S, Davies WG (2020). Heart Failure Drug Modifies the Intrinsic Dynamics of the Pre-Power Stroke State of Cardiac Myosin. Journal of Chemical Information and Modeling
10.1021/acs.jcim.0c00953
https://qmro.qmul.ac.uk/xmlui/handle/123456789/69421
Rehman S, Grigoryeva LS, Richardson KH, Corsini P, White RC, Shaw R, Portlock TJ, Dorgan B et al. (2020) . Structure and functional analysis of the Legionella pneumophila chitinase ChiA reveals a novel mechanism of metal-dependent mucin degradation. PLoS Pathogens vol. 16 (5)
10.1371/journal.ppat.1008342
https://qmro.qmul.ac.uk/xmlui/handle/123456789/64461
Tiberti M, Lechner BD, Fornili A (2019). Binding Pockets in Proteins Induced by Mechanical Stress. Journal of Chemical Theory and Computation vol. 15, (1) 1-6.
10.1021/acs.jctc.8b00755
https://qmro.qmul.ac.uk/xmlui/handle/123456789/54640
Hashem S, Tiberti M, Fornili A (2017) . Allosteric modulation of cardiac myosin dynamics by omecamtiv mecarbil . PLOS Computational Biology vol. 13, (11) e1005826-e1005826.
10.1371/journal.pcbi.1005826
https://qmro.qmul.ac.uk/xmlui/handle/123456789/28677
Tiberti M, Pandini A, Fraternali F, FORNILI A (2017) . In silico identification of rescue sites by double force scanning. Bioinformatics
10.1093/bioinformatics/btx515
https://qmro.qmul.ac.uk/xmlui/handle/123456789/25449
Pandini A, Fornili A (2016). Using Local States to Drive the Sampling of Global Conformations in Proteins . Journal of Chemical Theory and Computation vol. 12, (3) 1368 - 1379 .
10.1021/acs.jctc.5b00992
https://qmro.qmul.ac.uk/xmlui/handle/123456789/28561
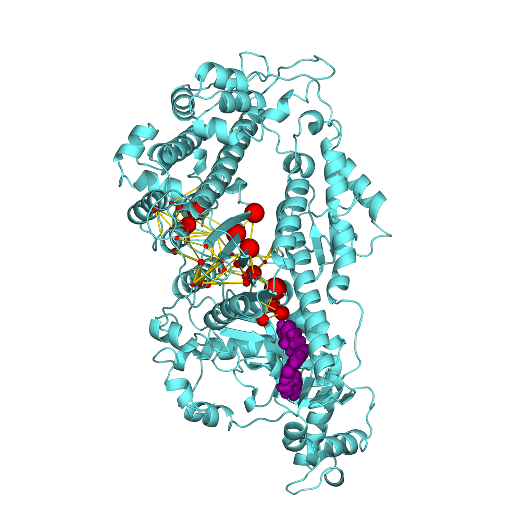
From: S. Hashem, M. Tiberti, A. Fornili (2017). Allosteric modulation of cardiac myosin dynamics by omecamtiv mecarbil. PLoS Computational Biology 13: e1005826. 10.1371/journal.pcbi.1005826
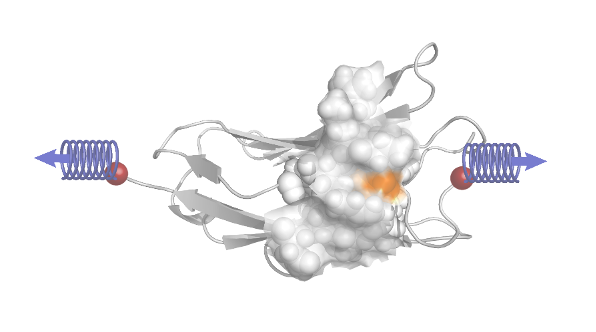
From: M. Tiberti, B.-D. Lechner, A. Fornili (2019). Binding Pockets in Proteins Induced by Mechanical Stress. Journal of Chemical Theory and Computation. 10.1021/acs.jctc.8b00755